First detailed look at how a molecular Ferris wheel delivers protons to cellular factories
No human cell can function without these tiny machines, which cause disease when they go haywire and offer potential targets for therapeutic drugs.
By Glennda Chui
All cells with nuclei, from yeast to humans, are organized like cities, with a variety of small compartments – organelles – that serve as factories where various types of work are done. Some of those factories, like the ones that break down and recycle molecules, need to continually pump in protons – hydrogen atoms with their electrons stripped off – to maintain the acidic environment they need to do their job. For this they rely on molecular Ferris wheels.
Embedded in the organelle’s fatty outer membrane, these microscopic machines have rotors that spin 100 times per second, picking up protons from outside the organelle and dropping them off on the inside.
Now scientists have figured out a key step in how these Ferris wheels work in a yeast proton pump known as vacuolar ATPase (V-ATPase). The results of their study, which combined high-resolution images made at the Department of Energy’s SLAC National Accelerator Laboratory with supercomputer simulations, were published in Science Advances today, giving scientists insight into a fundamental process that could potentially be harnessed to thwart disease.
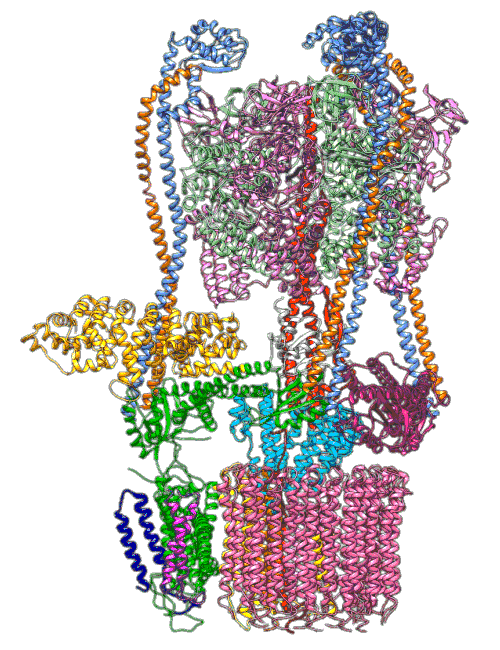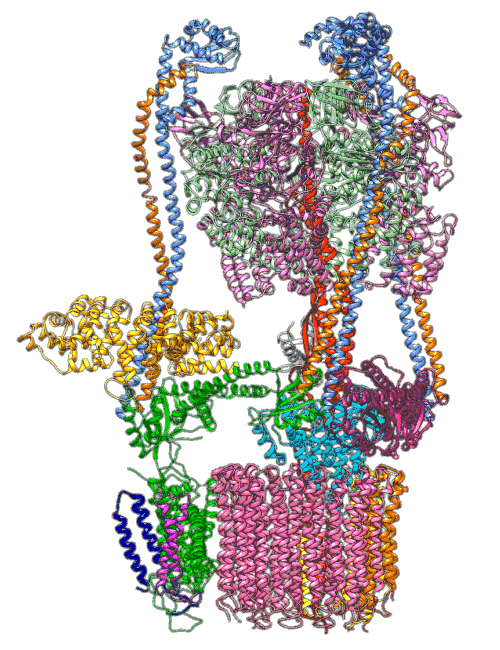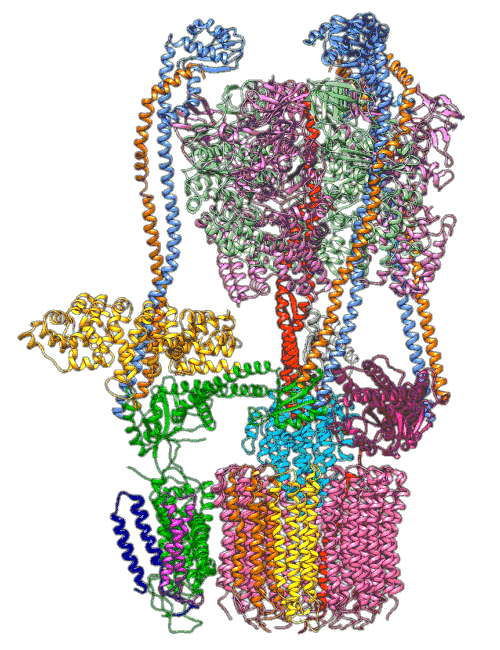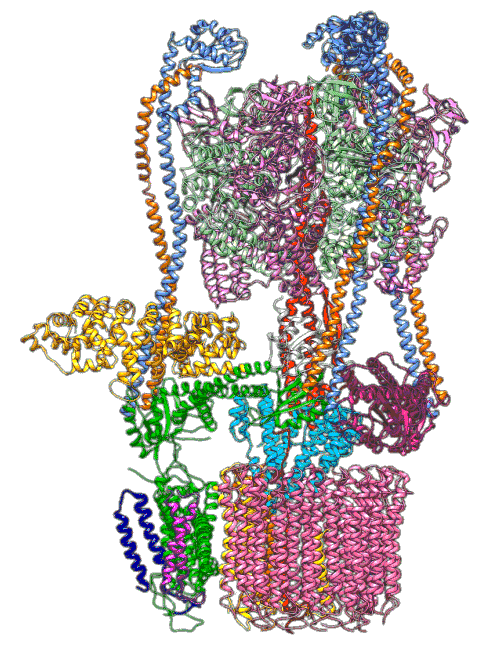
“The V-ATPase proton pumps perform a wide range of functions, from helping transmit nerve signals to helping specialized cells secrete acid for maintaining bone,” said Stephan Wilkens, a biochemist at SUNY Upstate Medical University and study co-author. “Malfunctions in these molecular machines contribute to diseases such as osteoporosis, neurodegeneration, diabetes, cancer and AIDS, so understanding them is important for human health.”
Wah Chiu, a professor at SLAC and Stanford and co-director of the Stanford-SLAC Cryo-EM Facilities where the imaging was done, said scientists are already investigating how these pumps in human cells might affect replication of the COVID-19-causing virus in patients. “It turns out the majority of therapeutic drugs on the market target molecular machines like this one that sit in cell membranes,” he added.
Watching the wheel go round
No human cell can function without proton pumps, which among other things help organelles intercept viruses and other pathogens and divert them to cellular trash bins.
While previous studies had determined the molecular structure and basic function of V-ATPases in a number of organisms, Wilkens said, “the big question was how do they work? To explain the mechanism it’s helpful to see it in action, just like the first serial snapshots of a galloping horse finally settled the question of whether it always had at least one hoof on the ground. The answer was no.”
In earlier cryo-EM research, Chiu, Wilkens, SLAC/ Stanford postdoctoral researcher Soung-Hun Roh and others produced high-resolution images that allowed them to identify the 10 amino acid “seats” on the yeast Ferris wheel that bind protons and carry them through the membrane to the organelle’s interior, as well as other amino acids that catch them when they arrive. Based on that picture, they suggested that the proton drop-off might be aided by water molecules, but their images were not sharp enough to confirm that the water molecules were there.
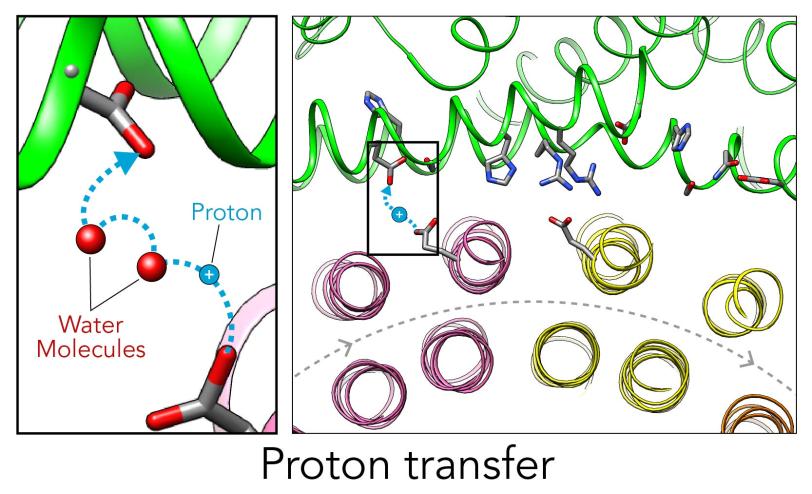
In the current study, thanks to another round of even higher resolution cryo-EM imaging at SLAC they were able to locate the water molecules around the suspected proton path. To make the proton pump motor come to life, a research group led by Abhishek Singharoy at the Arizona State University Biodesign Institute developed computer simulations of the process and ran them on a DOE supercomputer at Oak Ridge National Laboratory.
The simulations, which incorporated cryo-EM structures derived from images of the yeast Ferris wheel captured at two different points in its rotation, confirmed the experimentally observed water molecules lining up to form “wires” at the proton drop-off point. These wires convey protons from their seats on the Ferris wheel to landing spots inside the organelle, like a fire brigade passing buckets hand to hand, bridging a gap they couldn’t navigate on their own.
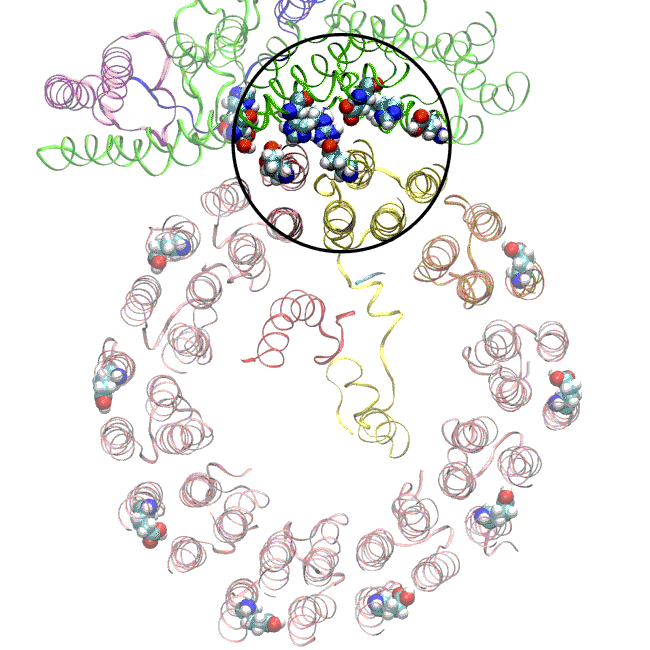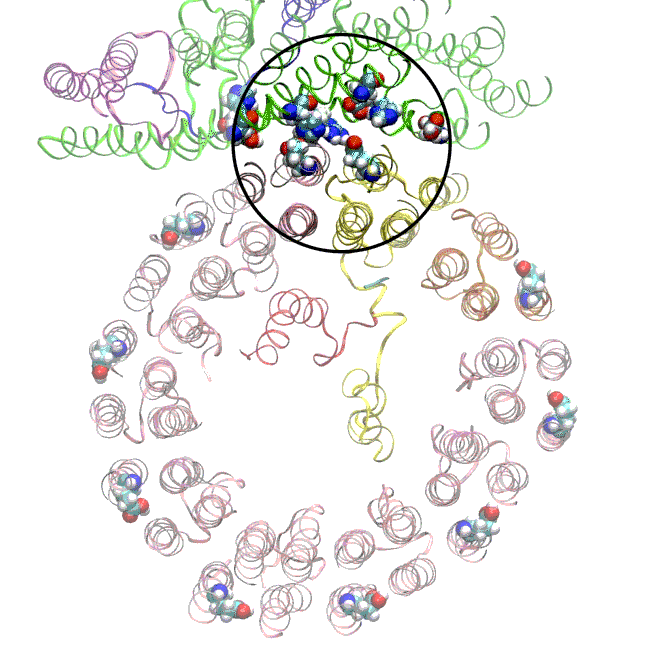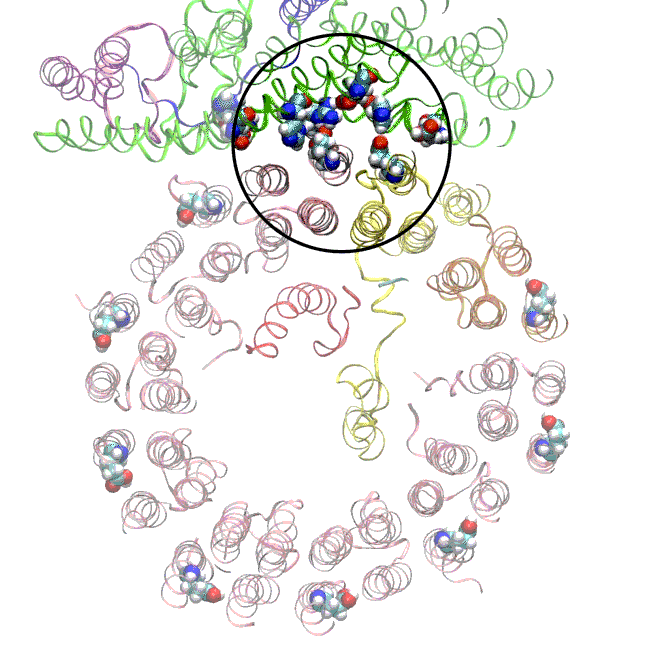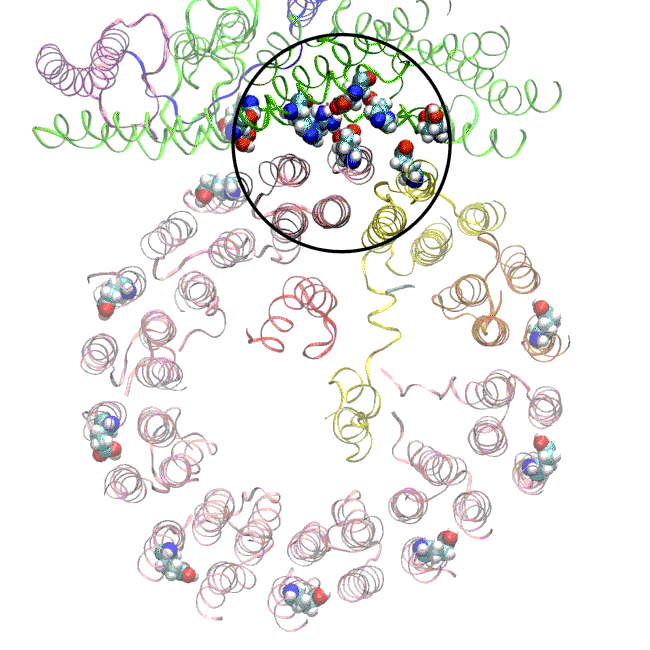
The simulations and the images matched nicely, Wilkens said, providing strong evidence that the picture they painted of the proton drop-off is correct.
Going forward, Chiu said, recent advances in cryo-EM that allow imaging of individual particles at atomic resolution – even when they take slightly different shapes – will open new opportunities for using it as a tool to discover effective drugs for illnesses involving proton pumps.
Soung-Hun Roh is now an assistant professor at Seoul National University. Scientists from the University of Lorraine in France and the University of Illinois at Urbana-Champaign also contributed to this research, which was funded by the U.S. National Institutes of Health and the Korean National Research Foundation. The Oak Ridge Leadership Computing Facility is supported by the DOE Office of Science.
Citation: Soung-Hun Roh et al., Science Advances, 7 October 2020 (10.1126/sciadv.abb9605)
Contact
For questions or comments, contact the SLAC Office of Communications at communications@slac.stanford.edu.
SLAC is a vibrant multiprogram laboratory that explores how the universe works at the biggest, smallest and fastest scales and invents powerful tools used by scientists around the globe. With research spanning particle physics, astrophysics and cosmology, materials, chemistry, bio- and energy sciences and scientific computing, we help solve real-world problems and advance the interests of the nation.
SLAC is operated by Stanford University for the U.S. Department of Energy’s Office of Science. The Office of Science is the single largest supporter of basic research in the physical sciences.






